Research overview
We investigate how sequence information in biological macromolecules governs recognition, binding, and dynamical structure. We use and develop cutting edge techniques for doing high-throughput measurements of molecular binding, which we use to build quantitative models of how life works at the molecular level.
From sequence, to structure, to function in macromolecular binding and recogntion
Perhaps the best-studied phenomena in molecular biology is the manner in which information inside a cell flows – i.e. how genetic information is stored and transformed into functional proteins. However, DNA, RNA and protein sequences encode complementary layers of information, equally important for the cell function, yet far less fully understood. This “hidden” information encodes specific recognition sequences for the binding of DNA, RNA and protein by other regulatory RNAs and proteins, essential for regulating cellular processes in all lifeforms. In general, we are currently unable to quantitatively predict molecular recognition, dynamical structure and function from a given nucleotide or protein sequence. Gaining this quantitative understanding is of general interest in all of molecular biology, and of special interest for transcription factor (TF) - DNA binding. This binding and recognition control gene expression, and can cause a myriad of disease states when the binding goes wrong.
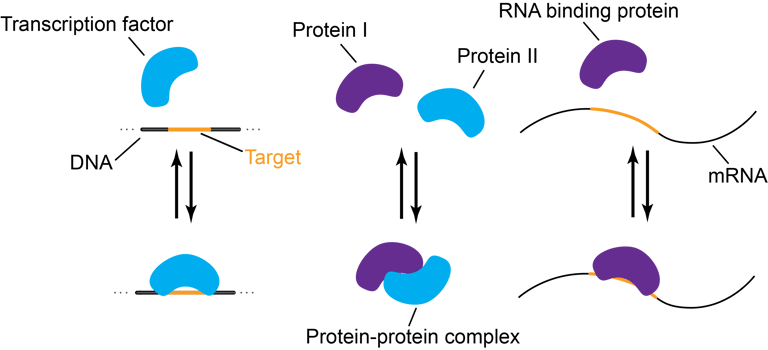

The primary sequence of biomolecules determines who binds who.
Parallelized, simultaneous binding measurements on diverse sequence mutants
To understand the sequence dependence on macromolecular binding, we use and develop cutting edge techniques for doing high-throughput measurements of molecular recognition. In these experiments we probe biophysical parameters of thousands to millions of sequence variants in a single measurement.
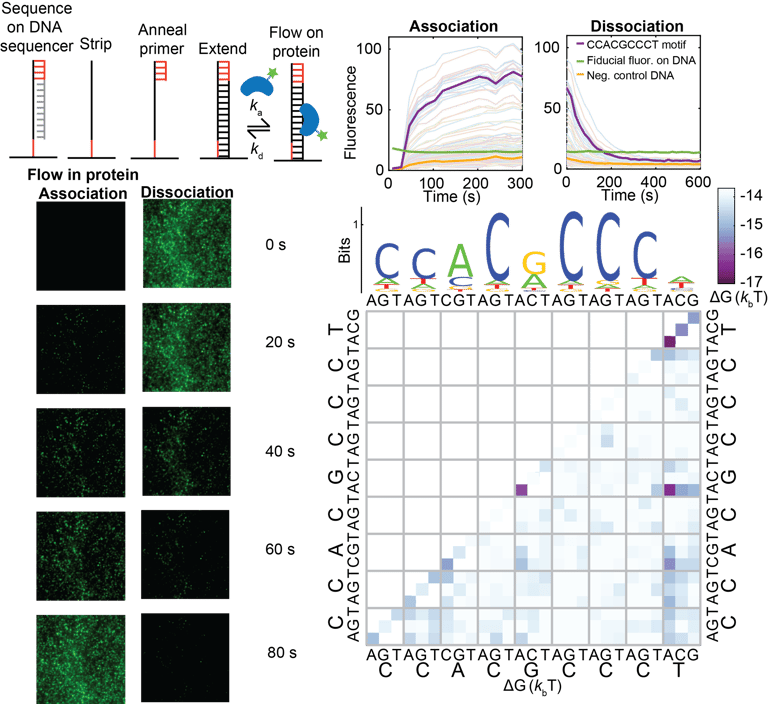
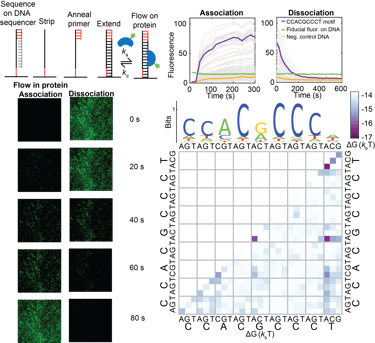
Measuring the kinetics and equilibrium binding of a transcription factor to many DNA binding site mutants at the same time.
Building quantitative models for prediction and mechanistic understanding
When the binding data has been gathered, we use it to build quantitative models of how protein and nucleotide sequence determines binding specificity. These models can be both predictive, like deep learning models, or mechanistic, giving us novel biological insights on how these interactions actually work on the molecular level. Previously, we have determined how DNA binding proteins manage to find their specific target sites in the genome without getting stuck too long at false targets. We found that the binding strength is determined by how fast the target is recognized - and not how long the protein remains in the bound conformation. This is the opposite to how sequence specific affinity is currently explained in textbooks and undergraduate molecular biology courses.
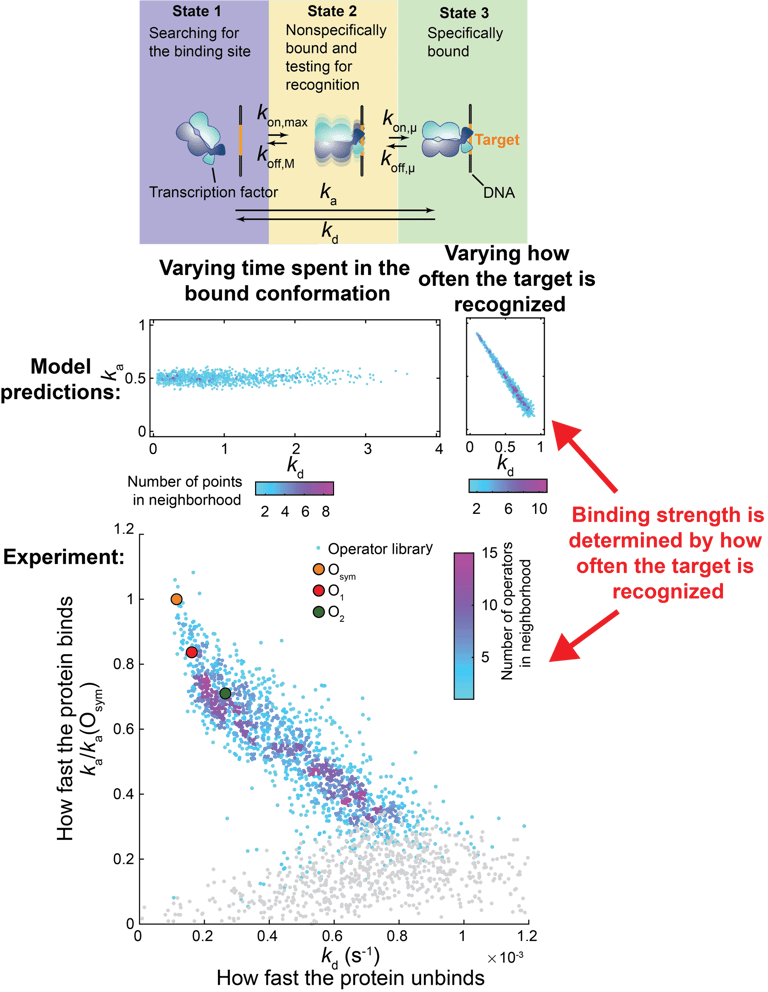
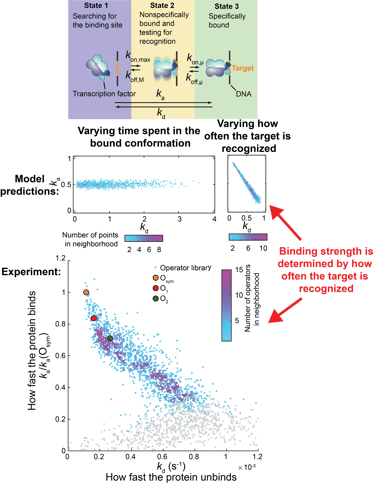
Binding strength is determined by how often the target is recognized.
